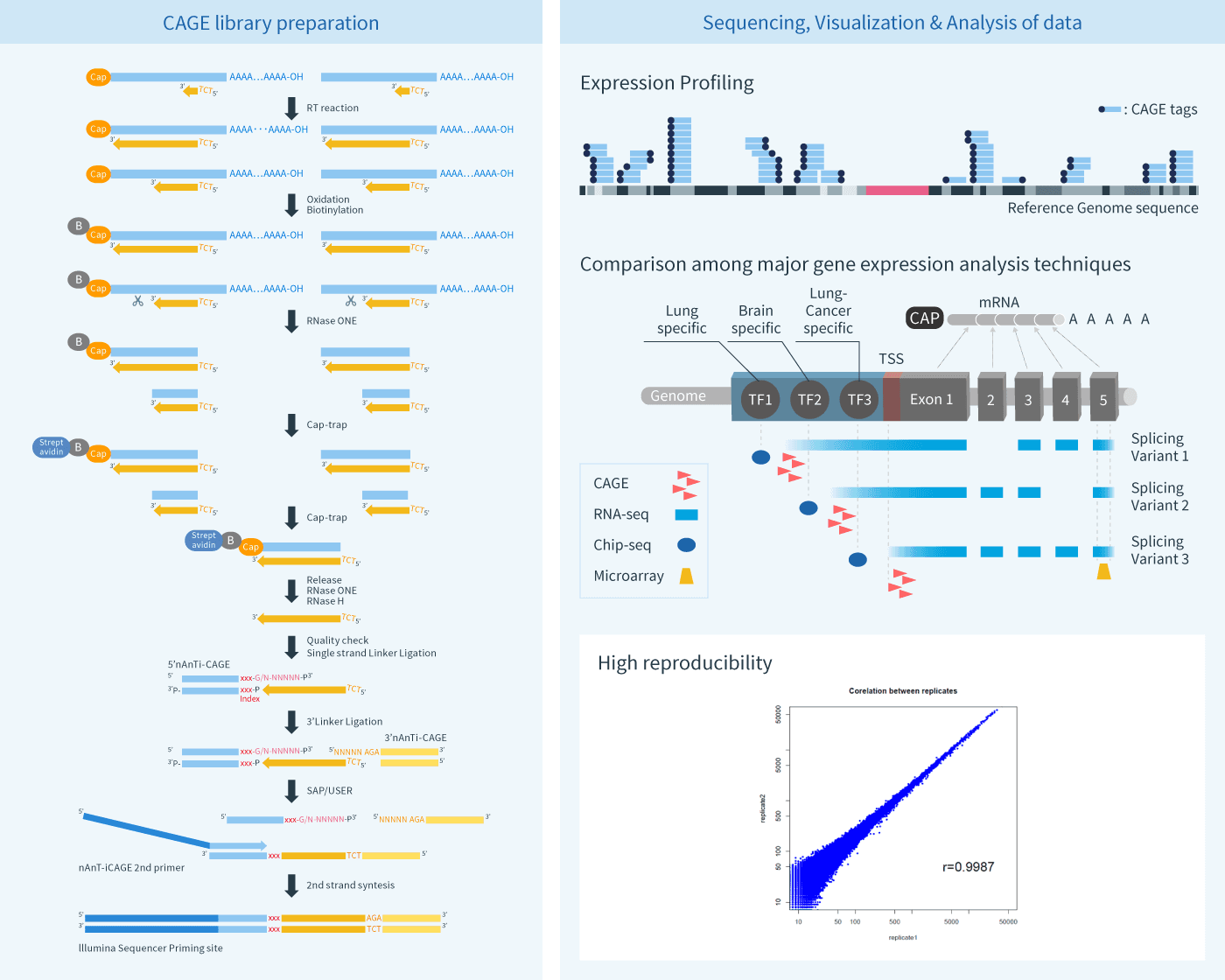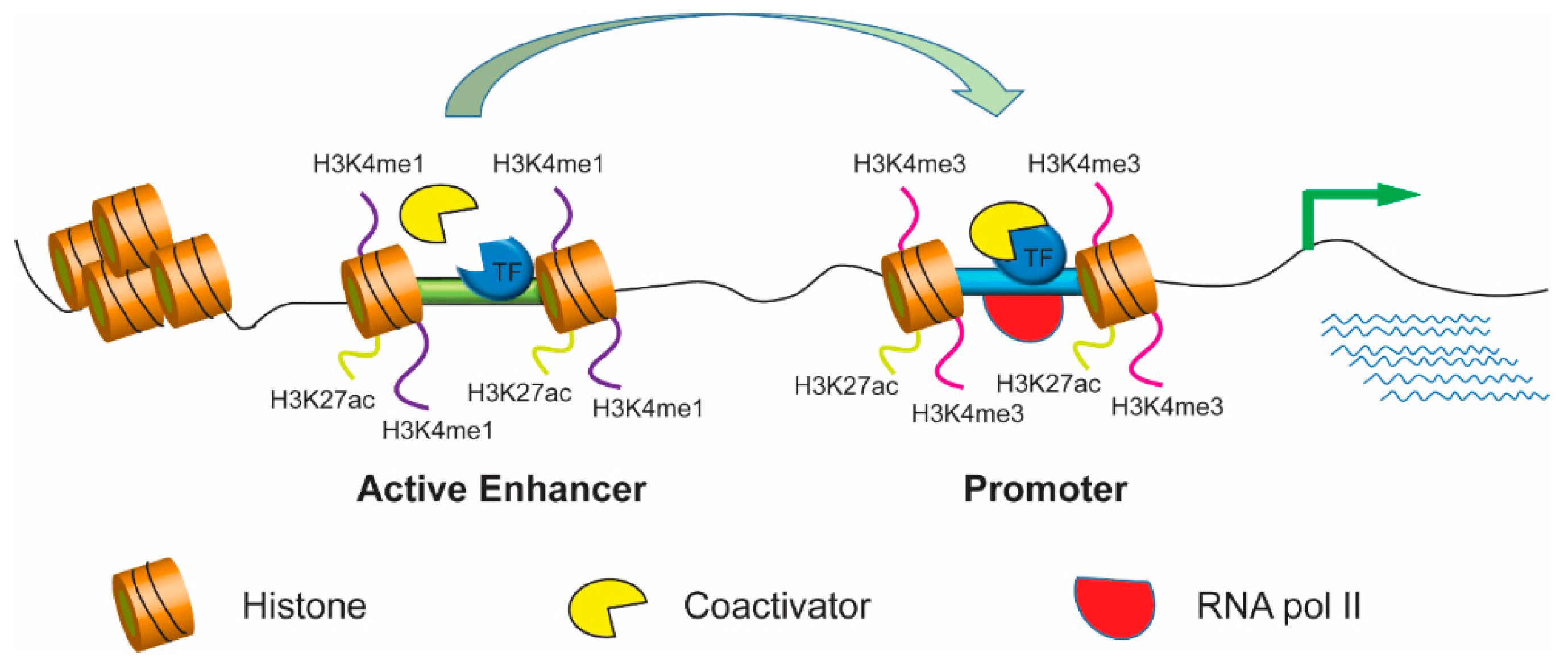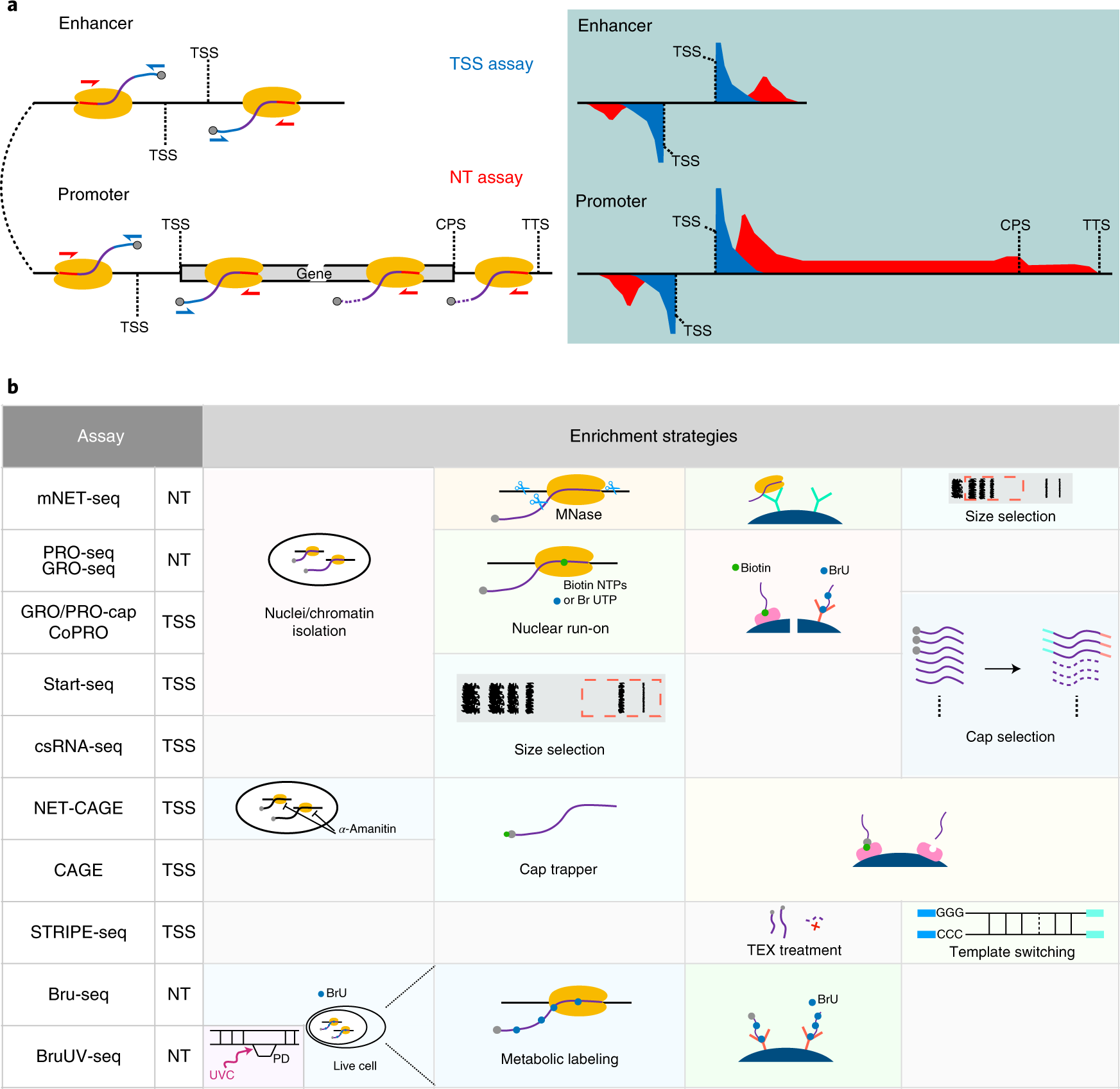
A comparison of experimental assays and analytical methods for genome-wide identification of active enhancers | Nature Biotechnology

Enhancer and super‐enhancer: Positive regulators in gene transcription - Peng - 2018 - Animal Models and Experimental Medicine - Wiley Online Library

Landscape of Enhancer-Enhancer Cooperative Regulation during Human Cardiac Commitment: Molecular Therapy - Nucleic Acids

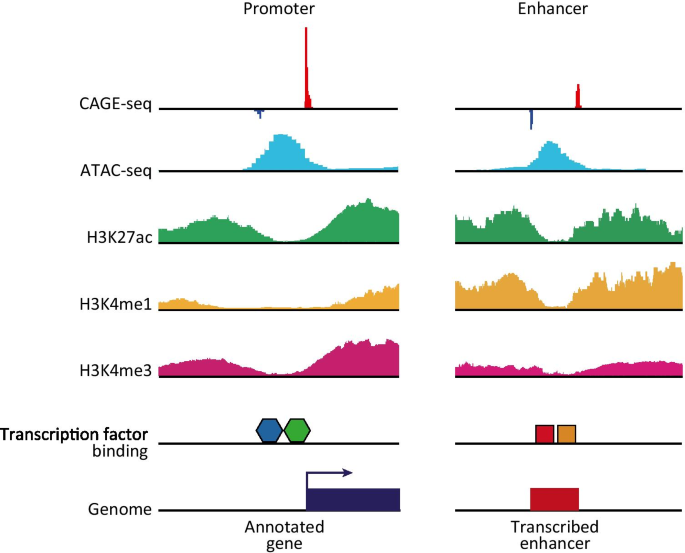
De novo identification of enhancers with higher sensitivity in NET-CAGE... | Download Scientific Diagram
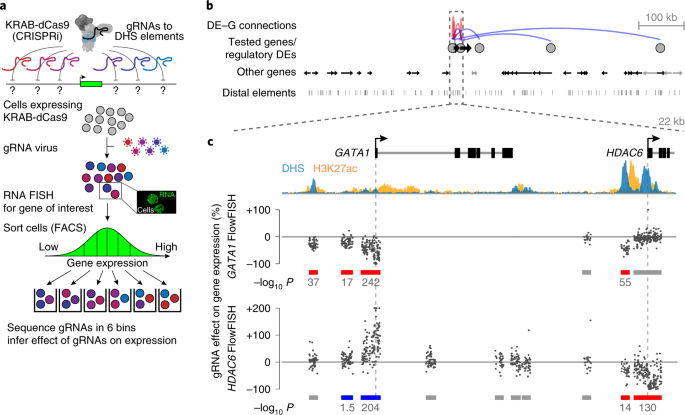
Activity-by-contact model of enhancer–promoter regulation from thousands of CRISPR perturbations | Nature Genetics
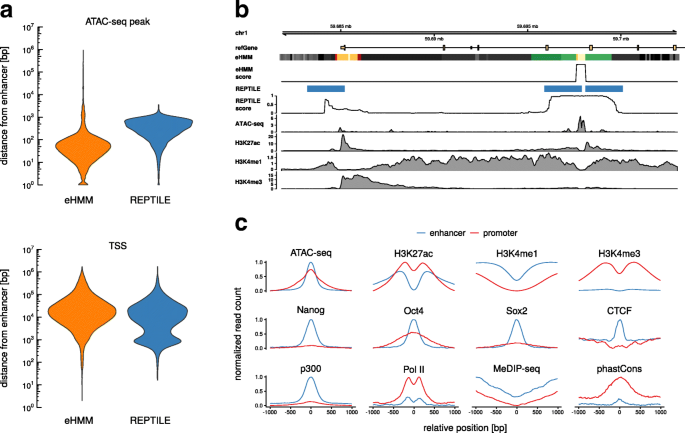
Predicting enhancers in mammalian genomes using supervised hidden Markov models | BMC Bioinformatics | Full Text

A neural network based model effectively predicts enhancers from clinical ATAC-seq samples | Scientific Reports
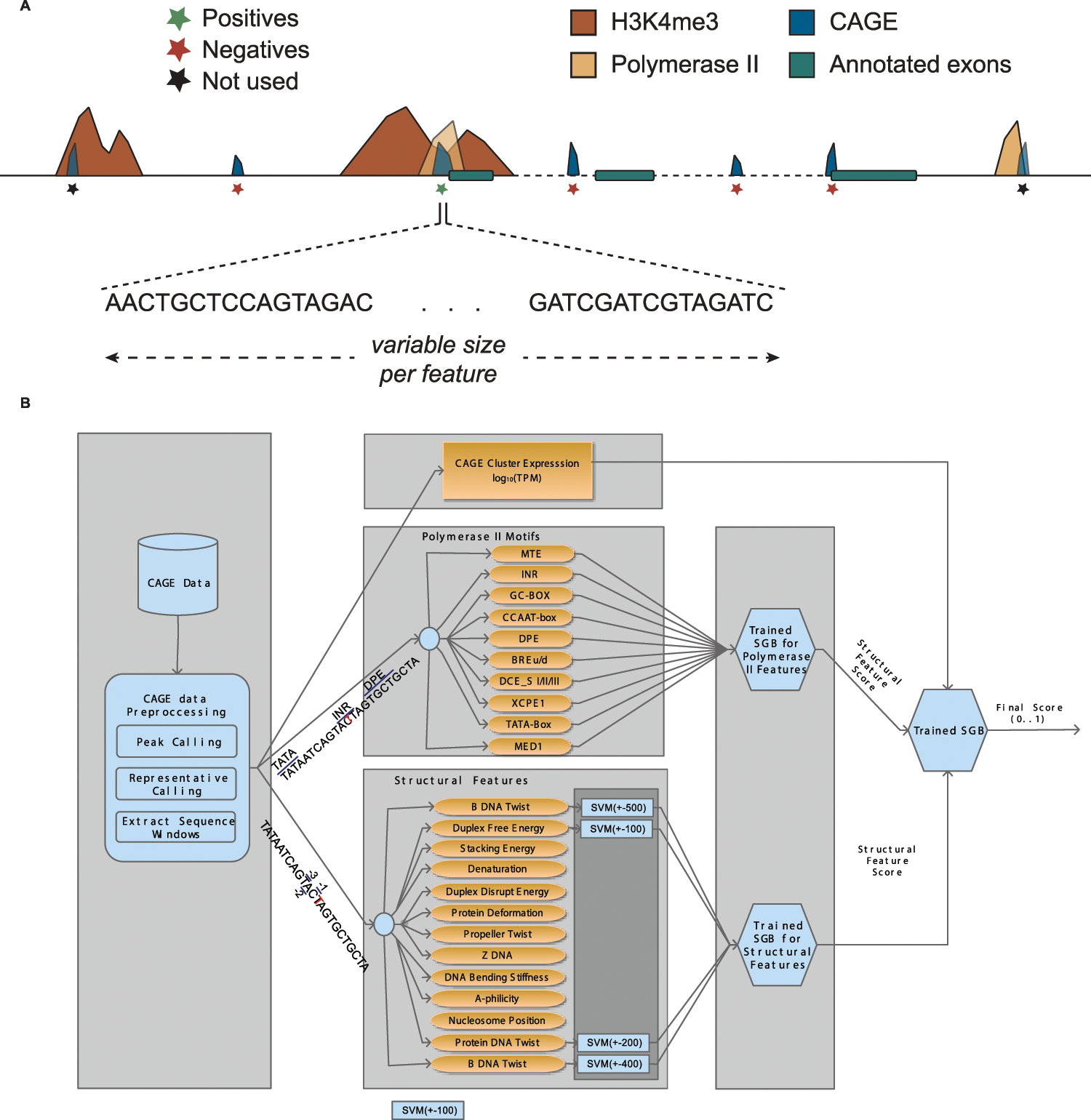
Solving the transcription start site identification problem with ADAPT-CAGE: a Machine Learning algorithm for the analysis of CAGE data | Scientific Reports

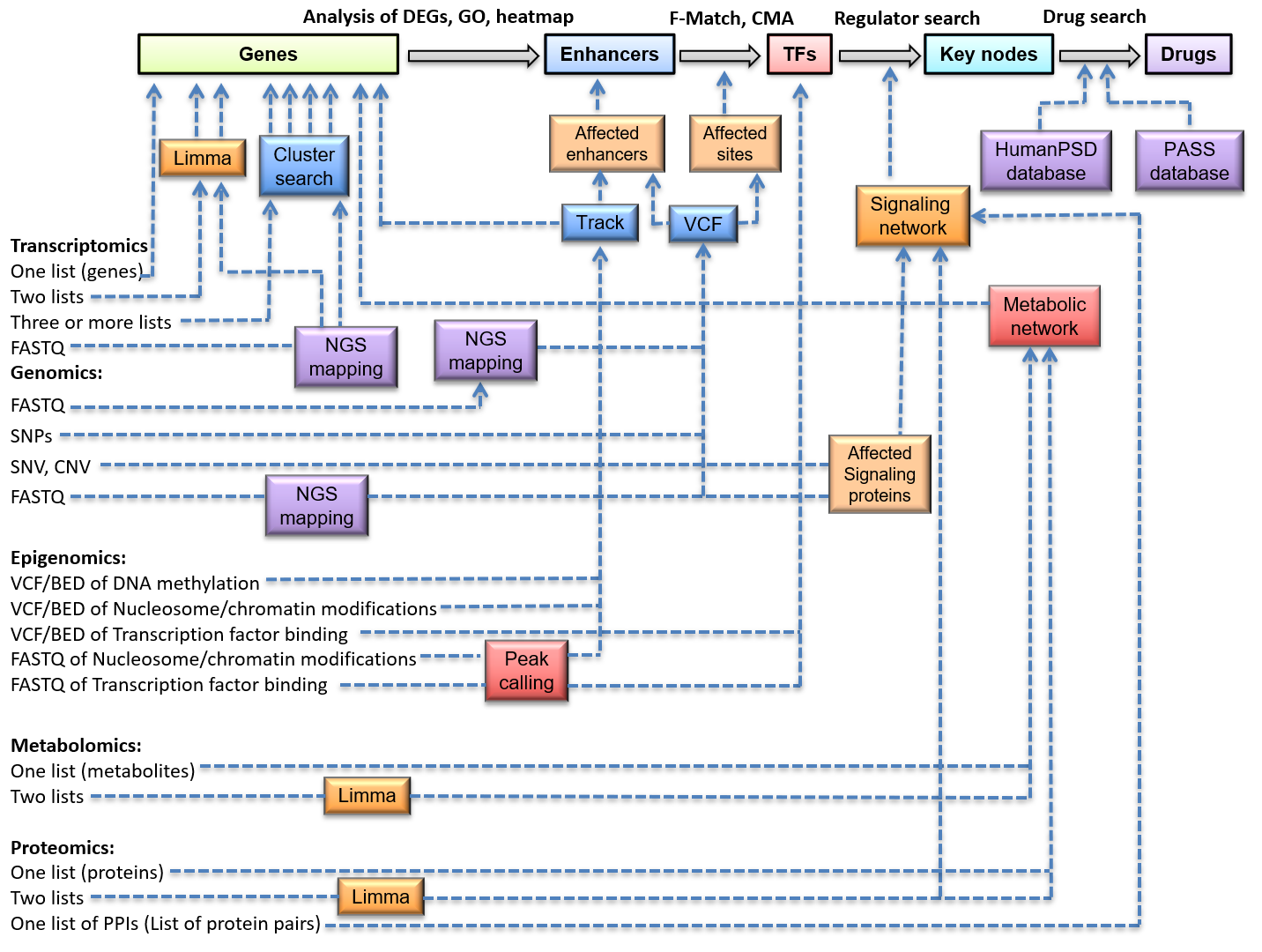
Enhanced Identification of Transcriptional Enhancers Provides Mechanistic Insights into Diseases: Trends in Genetics

De novo identification of transcribed enhancers and super-enhancers in... | Download Scientific Diagram

A High-Resolution Map of Human Enhancer RNA Loci Characterizes Super- enhancer Activities in Cancer - ScienceDirect
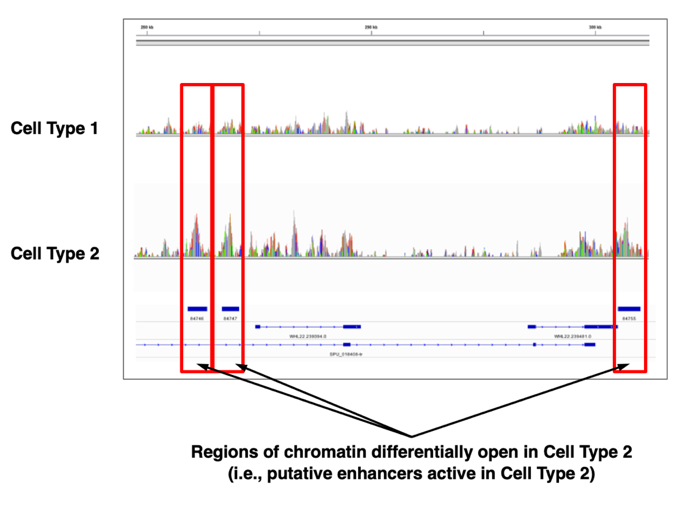
Genomic Feature Datasets for Analysis of Regulatory DNA Elements – Resource for Developmental Regulatory Genomics
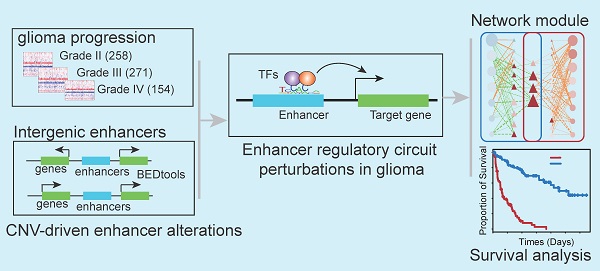
Systematic analysis of enhancer regulatory circuit perturbation driven by copy number variations in malignant glioma

Altered Enhancer and Promoter Usage Leads to Differential Gene Expression in the Normal and Failed Human Heart | Circulation: Heart Failure

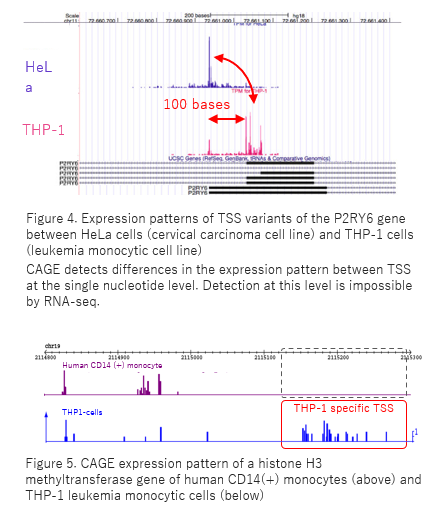
Frontiers | Mapping Mammalian Cell-type-specific Transcriptional Regulatory Networks Using KD-CAGE and ChIP-seq Data in the TC-YIK Cell Line

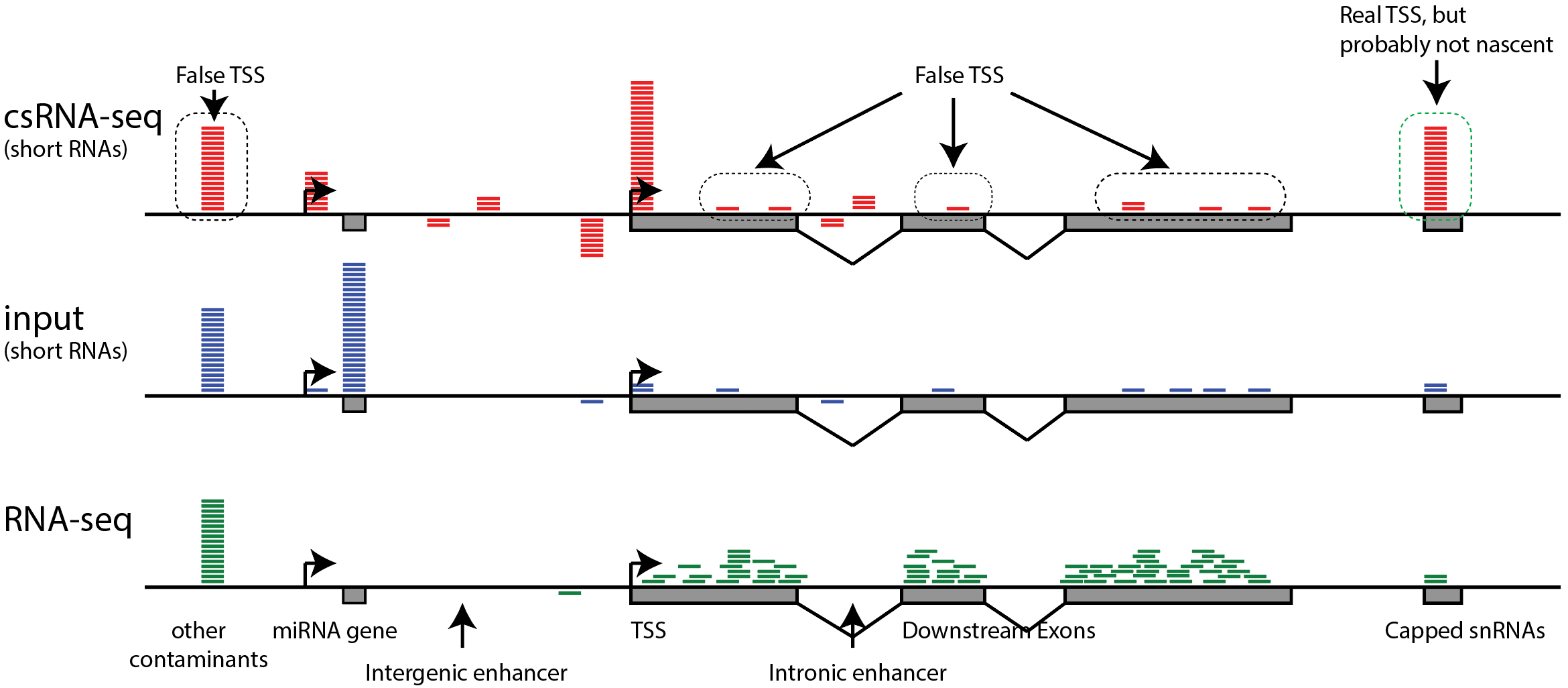
Nascent RNA sequencing analysis provides insights into enhancer-mediated gene regulation | BMC Genomics | Full Text